Please contact us for any questions or request for reagents for this structure. |
HIP2 - Human Huntingtin Interacting Protein 2 (aka E2-25) |
|
|
Huntington's disease is an autosomal dominant neurodegenerative disorder characterised by death of cortical and striatal neurons and caused by an expanded polyglutamine tract at the amino terminus of the protein huntingtin. Although the pathogenic mechanism is still unknown, important disease phenotypes have been discovered, such as neuronal intranuclear inclusions of mutant huntingtin and the presence of specific aggregate-interacting proteins. Two important huntingtin-associated proteins are ubiquitin and, via yeast-two-hybrid experiments and in-vitro pull-downs, the ubiquitin-conjugating enzyme E2-25, also known as huntingtin interacting protein 2 (HIP2). Huntingtin is an extremely large protein of about 3000 amino acids, widely and ubiquitously expressed, and part of a protein network that regulates membrane trafficking. |
|
|
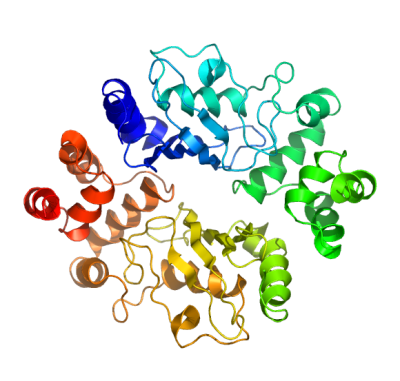
HIP2 presumably contributes to the ubiquitination of huntingtin, possibly through an E3-ligase independant and dependent manner. We have determined the high resolution structure of HIP2. In addition to the canonical ubiquitin conjugating (UBC) fold, our data reveals the structure and orientation of the carboxy-terminal ubiquitin associated domain (UBA) (residues 159-194). This helical bundle forms extensive interactions with the alpha-3 helix (139-153) of the core UBC domain; aromatic residues (residues 80,148,152,162,188) from both the UBC and UBA dominate at the interface. The UBA subdomain is spatially removed from both the reactive cysteine (Cys92) and from the putative E3-ligase binding surface (residues 4,8,11,67,68,102,103). The ubiquitin associated domain, which in other proteins has been shown to function as a ubiquitin binding module, may provide an additional site for ubiquitin binding, potentially directing specificity for polyubiquitylation reactions. |
|
