Workshop - iSee: Interactive 3D Documents for the Dissemination of Structural Biology
May 10th, 2011 - Toronto, Canada
|

|
This workshop will be limited to 30 (thirty) participants.
|
Do you wish that interactive molecular graphics could be integrated as part of your papers or documents, making your work more accessible to non-structural biologists? Or how about reading a document that allowed you to manipulate an embedded 3D visualisation of a model without having to resort to loading and trying to reproduce the figure just to grasp what the authors wanted to say?
The SGC, in collaboration with MolSoft LLC (http://www.molsoft.com) have developed the iSee concept [1-3; www.thesgc.org/iSee and www.whatisisee.org ] for the dissemination of structural biology in interactive 3D documents. iSee is a free and platform-independent allowing anyone to read, author and share an interactive 3D document via the web or via Microsoft Office applications on Windows such as PowerPoint.
The iSee concept has been successfully used in academia for teaching and presentations and by the academic research community to disseminate and exchange structural information across different departments (e.g. SGC iSee datapacks, several pharmaceutical companies). More recently, it has begun to be used in a number of peer-reviewed journals such as PLoS Biology, PLoS ONE, Nature: Structural & Molecular Biology and Molecular & Cellular Proteomics, with other publishers in the works.
In this workshop we will demonstrate how the iSee platform can be used to create, share and prepare interactive 3D structural biology documents for presentation, peer-reviewed publication or dissemination on your own website.
|

|
Requirements for the workshop:
|
- Basic understanding of structural biology, structure coordinates (PDB) and computer graphics programs such as PyMol or Jmol.
- Your own laptop:
- OS: Windows XP/ Vista/ Windows 7, Mac OS X 10.6+, Linux (various flavours)
- 150 Mb of disk space
- 1Gb or more RAM
- A recent graphics chipset which supports hardware accelerated 3D (preferably not Intel, AMD/ATI or NVidia is preferred)
- Wi-Fi enabled
- We will be using the ‘Pro’ version of ICM for this workshop. A 4-weeks trial license will be available for the participants, who will be contacted after registration with instructions on how to proceed.
Note: If you have further question about the system requirements, please send an e-mail to wenhwa.lee@sgc.ox.ac.uk
|
Examples of iSee 3D documents: Click on the links below and follow the instructions.
|
- From the SGC iSee collection:
|
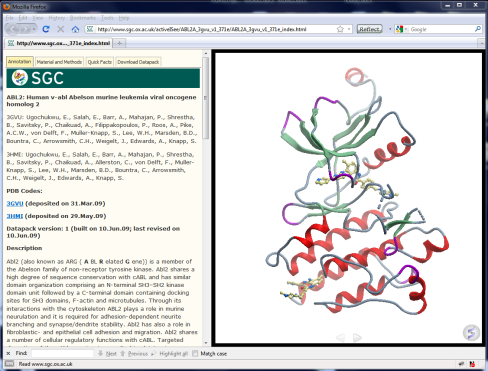
|
- From the PLoS ONE enhanced articles collection:
|
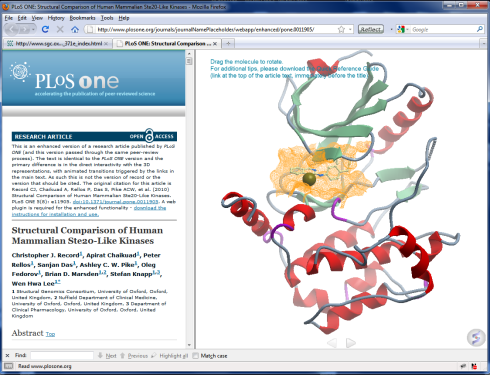
|
- From the Nature: Structural & Molecular Biology:
|
|
References
- Abagyan et al., TiBS 2006, 31(2):76-8
- Lee et al., PLoS One 2009, 4(10):e7675
- Raush et al., PLoS One. 2009, 4(10):e7394
|
|